Pinda - Pipeline for INtraspecies Duplication Analysis
NAME
Pinda -- Pipeline for Intraspecies Duplication Analysis
DESCRIPTION
A Web service aiming to facilitate detection of specific gene
duplications in an organism species of choice.
AVAILABILITY
Pinda is located at http://orion.mbg.duth.gr/Pinda, whereas the Source Code can be obtained at https://github.com/dgkontopoulos/Pinda.
USAGE
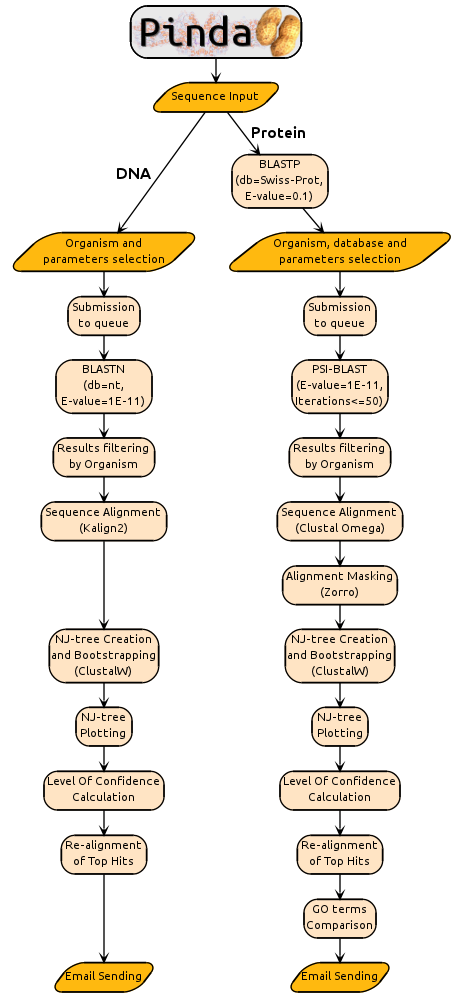
At Pinda's starting page, you may enter a Protein or DNA sequence. After clicking the "Continue" button, Pinda will take you to the next page, where you may choose the organism and the database within which the duplication analysis will take place. You also have to enter your email address, so that you can be notified after the job is complete. Past this point, your task has entered the queue. You will receive its results via email, including a possible duplications table, a multiple sequence alignment and a dendrogram.
FLOWCHART
AUTHOR
Pinda has been developed by Dimitrios - Georgios Kontopoulos under the supervision of Prof. Nicholas M. Glykos at the Department of Molecular Biology and Genetics of Democritus University of Thrace, Greece.
LICENSE
This program is free software: you can redistribute it and/or modify
it under the terms of the GNU Affero General Public License as
published by the Free Software Foundation, either version 3 of the
License, or (at your option) any later version.
For more information, see http://www.gnu.org/licenses/.

